VarSome's somatic variant classification explained
Charles Chapple, PhD, VarSome's Head of Bioinformatics, demonstrates the power of VarSome's automated implementation of AMP guidelines for cancer variant interpretation.
Learn about the new VarSome cancer classifier module, based on the joint recommended guidelines from the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists.
Charles Chapple, PhD, VarSome's Head of Bioinformatics, demonstrates the power of VarSome's automated implementation of AMP guidelines for cancer variant interpretation.
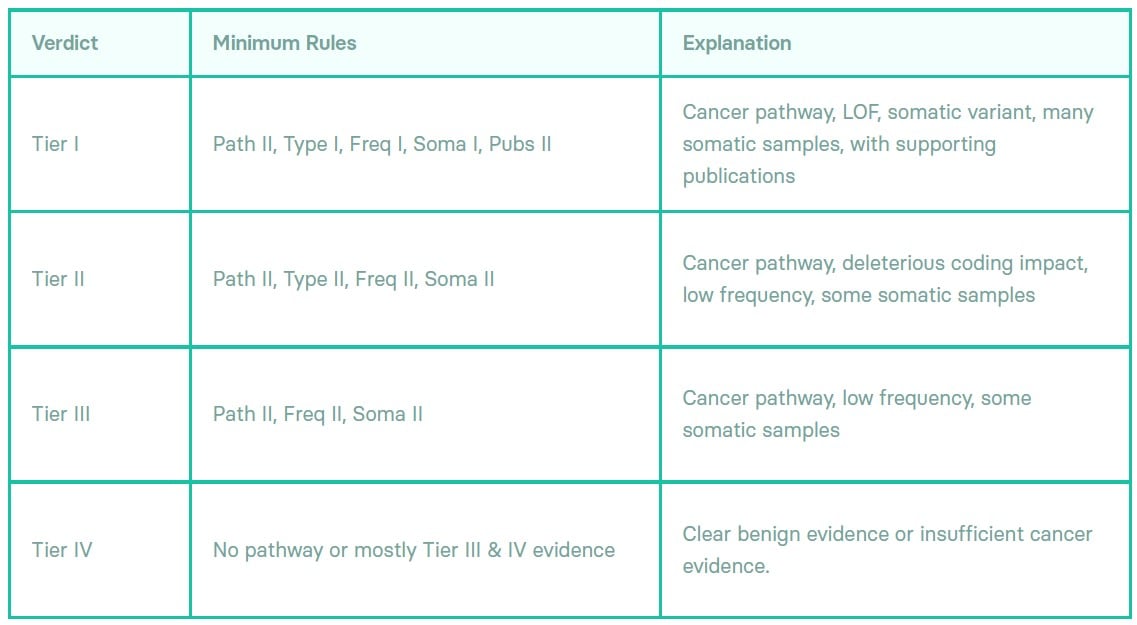
Our implementation is derived from the ”Standards and Guidelines for the Interpretation and Reporting of Sequence Variants in Cancer”, published as a joint recommendation in 2017 from the Association for Molecular Pathology, American Society of Clinical Oncology, and College of American Pathologists.
Our guiding principle throughout, following the advice of our clinical advisors, has been to implement a rigorous evidence-based approach to identifying potential cancer variants.
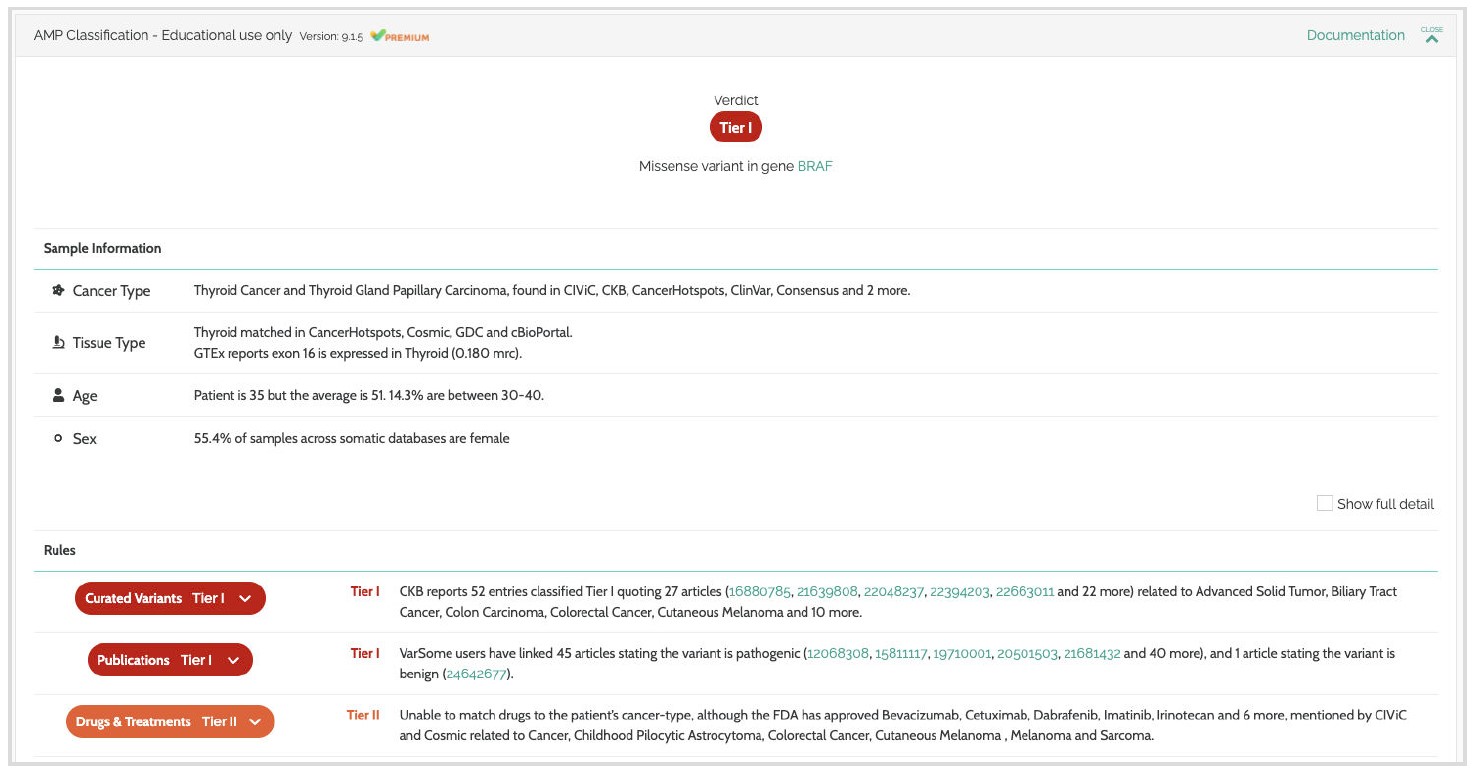
All the rules provide clear natural language explanations of why the were triggered and which evidence was used, or, conversely, a full explanation of why the criteria were not met (these 'negative' explanations are displayed if 'show full detail' is ticked, but they are not retained in the Clinical platform for Tier IV variants.
We have leveraged a wide range of public-domain and commercial cancer databases, therefore the end result will partially depend on the user's VarSome.com subscription level.
VarSome Suite is brought to you by Saphetor SA.
© 2024 Saphetor SA – All Rights Reserved.