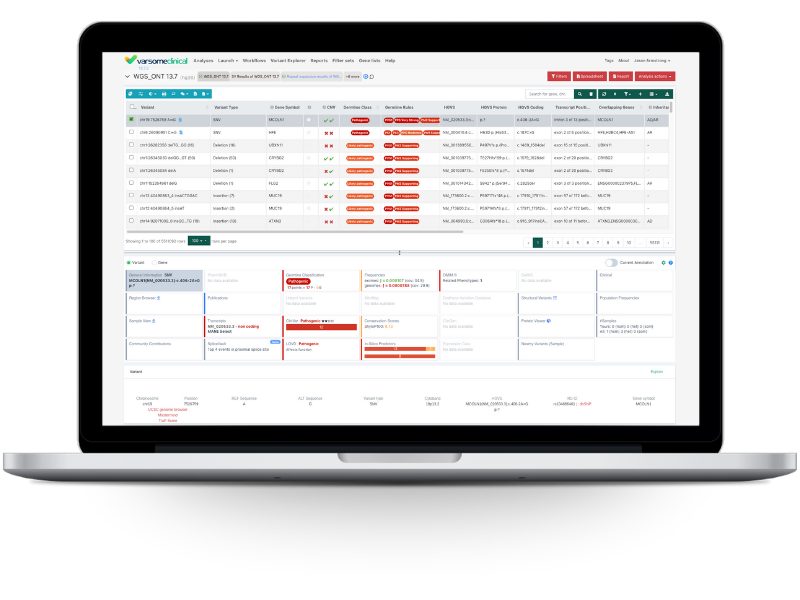
Identify key variants quickly with powerful, intuitive filtering capabilities.
Apply customizable, dynamic filters like allele frequency, pathogenicity, gene lists, and more. Make use of sophisticated algorithmic filters for complex analyses like compound heterozygous variants, de novo variants from trios, or carrier screening for couples. And prioritize causative variants efficiently with VarSome Picks, our phenotype-driven variant prioritization tool.
Our renowned classification engine supports both germline and somatic workflows, with modules based on ACMG/AMP guidelines.
Each classification is fully transparent, with guided criteria and clear audit trails. Users can adjust rule strength based on clinical or familial context, giving you control while maintaining consistency. Whether you’re working with inherited disorders or tumor samples, the system adapts to your interpretation framework.
Our Sample View provides a comprehensive visual inspection of all SNVs, CNVs, and structural variants in your samples, helping you to see your variants in context.
Perform multi-sample comparisons for scenarios like tumor-normal, trios, or large cohorts, including region overlap and compound heterozygous analysis. Build an internal sample database for frequency tracking and insights into variant recurrence across samples.
VarSome Clinical offers flexible deployment options and seamless integration capabilities to fit your bioinformatics workflow.
You can access VarSome Clinical as an online software application with cloud deployments in various regions, including Switzerland, the USA, and Saudi Arabia. VarSome Clinical also lets you build your own database of samples and variants, with the option to share selected variant data with trusted partner institutions.
For somatic analysis, VarSome Clinical integrates datasets such as COSMIC, OncoKB™, and CKB directly into variant interpretation. You also get access to our somatic classifier, based on AMP/ASCO/CAP guidelines.
It supports TMB/MSI biomarkers and provides visualization tools, such as IGV, for reviewing variants in context. Users can define custom gene panels per cancer type and gain insight into variant recurrence across multiple samples, helping you see patterns in tumour genomics.
Cloud Deployments

Hosted by Google Cloud

Hosted by Saphetor

Hosted by Google Cloud

Hosted by Oracle
Hear From Our Partners
Gencell Pharma, a leading genomics company in Latin America, provides advanced sequencing and molecular diagnostics. In this series of clips, their team share how VarSome Clinical supports accuracy in variant calling and interpretation, increases speed in data processing, and strengthens collaboration between organizations. Our partnership has helped Gencell to expand capacity, improve quality, and deliver reliable results to patients and clinicians across the region.
Upload Your Sample
Annotation & Classification

Interpretation Simplified

Our Variant Knowledge Base.

Join Our Global Community.

Expert Support, Real Humans.
Benchmarking of variant calling software for whole-exome sequencing using gold standard datasets - Wong et al. 2025.
Precision oncology platforms: practical strategies for genomic database utilization in cancer treatment - Gazola et al. 2024.
Exploring the Genetic Causality of Discordant Phenotypes in Familial Apparently Balanced Translocation Cases Using Whole Exome Sequencing - Aristidou et al. 2022.
Want to read more? Click here for the list of publications.
Memberships & Partnerships
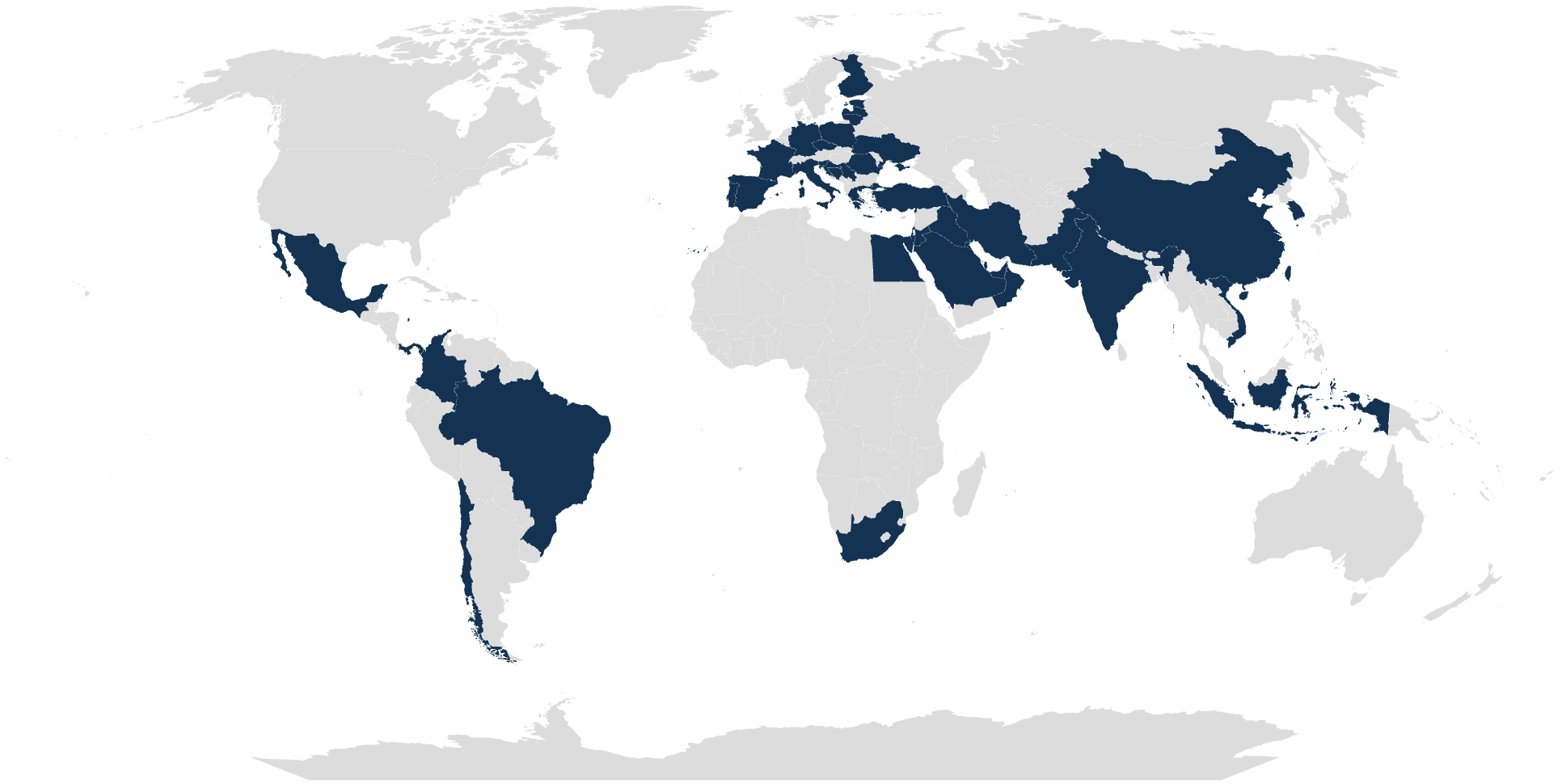
TRUSTED BY CLINICAL LABS WORLDWIDE




















VarSome Clinical is a Class C per Rule 3 of Annex VIII of the Regulation (EU) 2017/746 on in vitro diagnostic Medical Devices (IVDR).
VarSome Clinical is an in vitro diagnostic medical device software intended to qualitatively process high-throughput human DNA sequencing data to identify genetic variants, annotate them with relevant information from widely recognized databases, and report their classification verdict according to broadly recognized professional guidelines. VarSome Clinical is intended to be used by healthcare professionals to obtain information on the genomic status of the adult and pediatric population undergoing next-generation sequencing (NGS). The information obtained with VarSome Clinical may be used to support patient management decisions. VarSome Clinical does not provide any recommendations or conclusions related to diagnosis, prognosis, or treatment. The software is composed of two modules with distinct functions:
· VarSome Clinical - (Module 1) Variant Calling is intended for the qualitative analysis of high-throughput human DNA sequencing data, to identify discrepancies (variants) between DNA sequencing reads and the reference human genome. These discrepancies may represent genomic variants caused by single-nucleotide mutations, indels, and copy number variations. Inputs to Module 1 are next-generation FASTQ sequences that meet VarSome Clinical's quality requirements as depicted in the User Manual. The output of the Variant Calling module is a VCF text file format, which is the required input for VarSome Clinical Module 2.
· VarSome Clinical - (Module 2.1) Annotation is intended for automated annotation of genomic variants using relevant information from respected scientific sources. It assigns to the variants state-of-the-art functional information available from widely recognized databases cited within the User Manual. The level of annotated information is representative of the current understanding of the variant characterization, allowing for adequate application of variant classification rules (Module 2.2).
· VarSome Clinical - (Module 2.2) Variant Classification is intended for the classification of the annotated variants, based on semi-automated application of variant classification rules derived from broadly recognized professional guidelines, namely the guidelines published by the American College of Medical Genetics and Genomics (ACMG) and the Association for Molecular Pathology (AMP): o For germline variants, classification rules of the ACMG guidelines are followed, providing variants with one of the ACMG pathogenicity verdicts: "Benign", "Likely Benign", "Likely Pathogenic", "Pathogenic" or "VUS" (Variant of Unknown Significance). o For somatic variants, the classification rules of the AMP guidelines are followed, providing variants with one of the AMP pathogenicity verdicts: "Tier 1", "Tier 2", "Tier 3" or "Tier 4".
These verdicts are only reflective of the pathogenicity level as per state-of-the-art ACMG and AMP guidelines and do not directly provide information on the patient's clinical status. The semi-automated application of variant classification rules is intended as a standardized method for reaching an ACMG or AMP verdict, as per the ACMG and AMP guidelines cited in the User Manual, that reflects the current expert consensus at the time of the analysis. VarSome Clinical is indicated to qualitatively process high-throughput human DNA sequencing data to obtain information on the genomic status of the adult and pediatric population undergoing next-generation sequencing (NGS).
Read the User Manual before use.
Notified Body: TÜV SÜD Product Service GmbH
Manufacturer: Saphetor EPFL Innovation Park -C, CH-1015 Lausanne, Switzerland
EC Rep: Saphetor Life Sciences Europe, Leocharous 3, 10560 Athens Greece